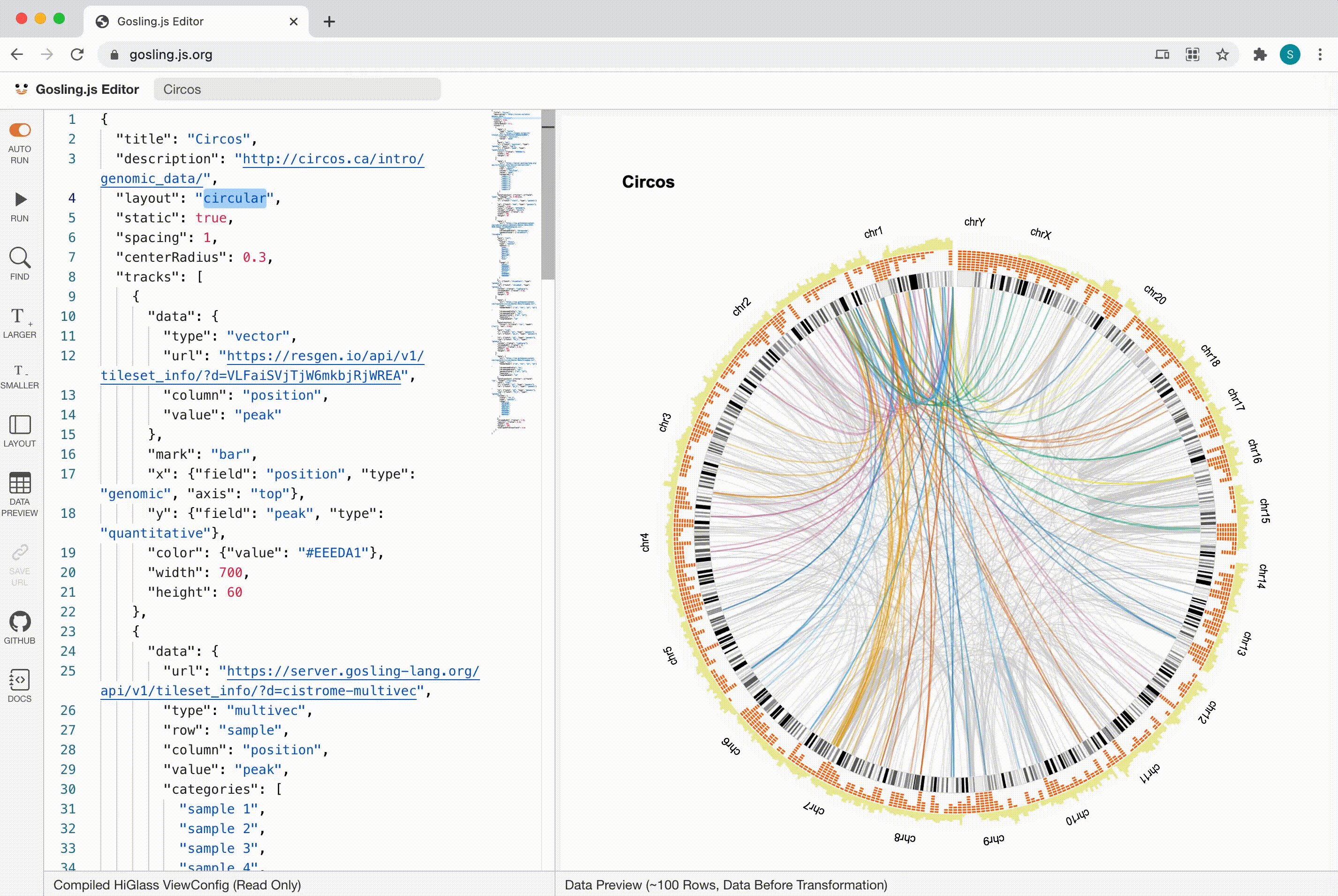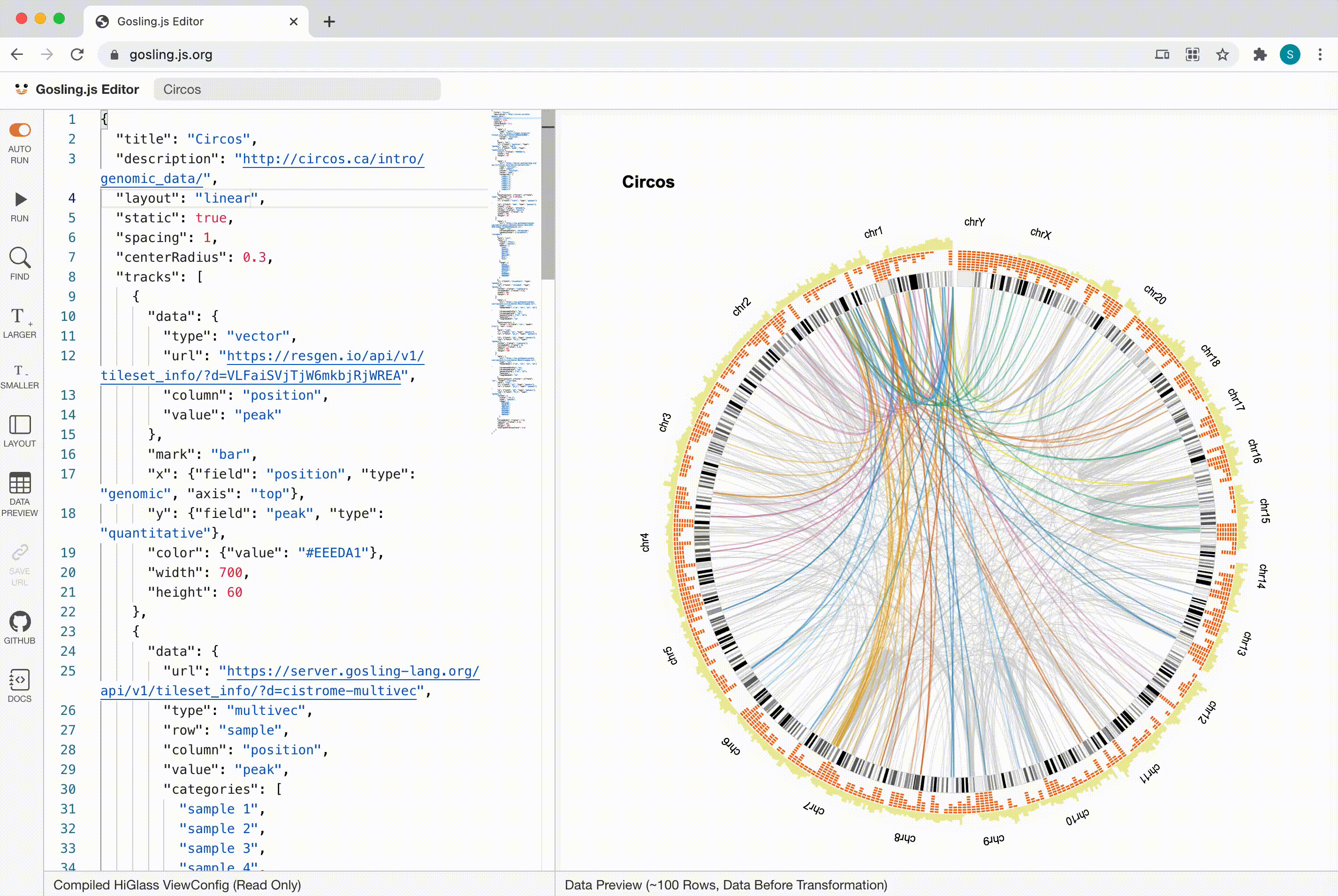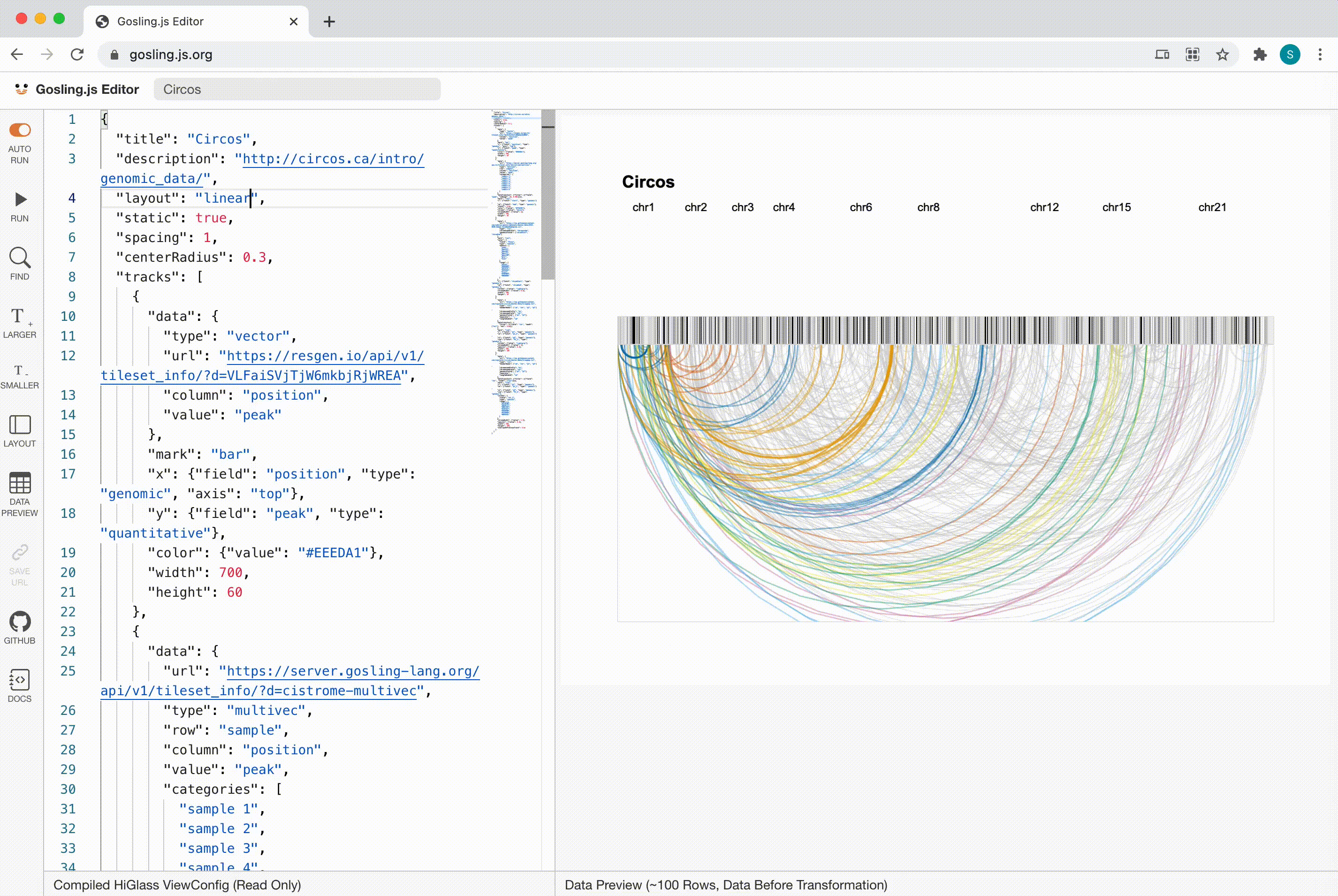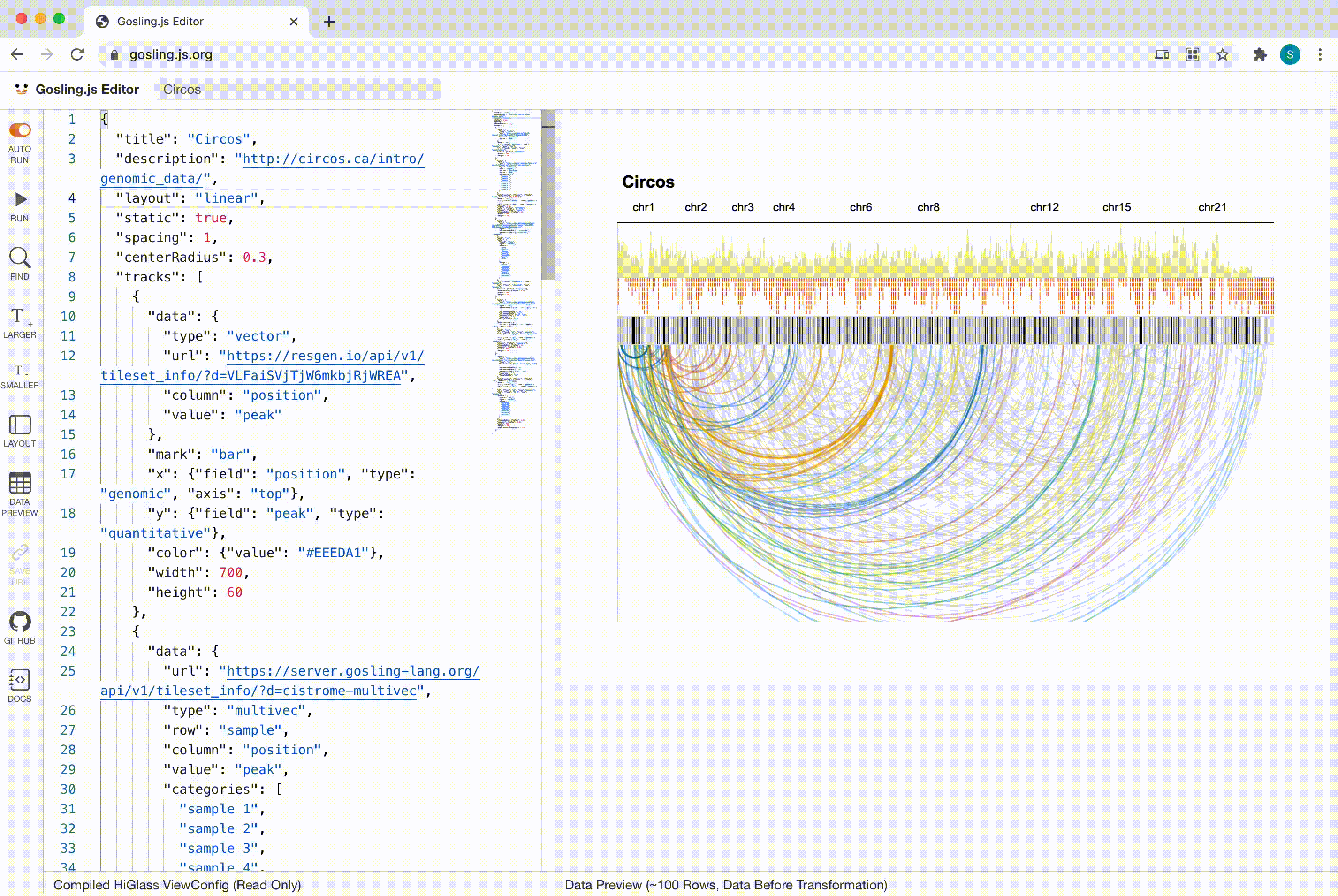
Gosling
Gosling
A Grammar-based Toolkit for Scalable and Interactive Genomics Data Visualization
Gosling.js: JavaScript library for GoslingGos: Python package for GoslingUse Gosling in your React AppUse Gosling in your Streamlit App
News
- We released Gosling v1.0.0! Read more in the announcement blog post.
- We release documentations for Gosling v0.10.0, which adds support for GFF files, a Javscript API to get the ID of views, and a placeholder track for non-Gosling visualizations.
- We release documentations for Gosling v0.9.30, which supports React v18 and BED file datafetcher!
- We release documentations for Gosling v0.9.20, which supports mouse event APIs,responsive design, and more expressive styles!
- Our tutorial on Gosling has been accepted for ISMB 2022 🎉
- Gosling won the Best Abstract Award at BioVis ISMB 2021 🏆
- Our first paper about Gosling.js has been presented at VIS 2021 and will be published to IEEE TVCG.
Scalability
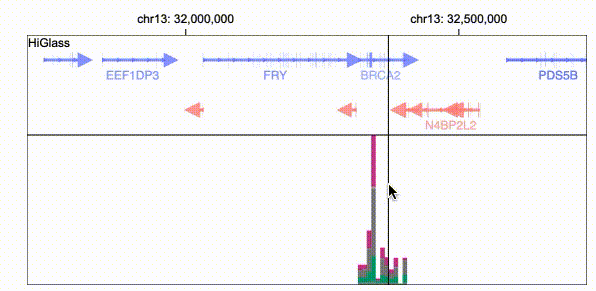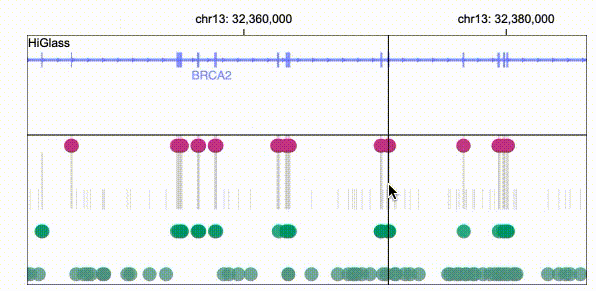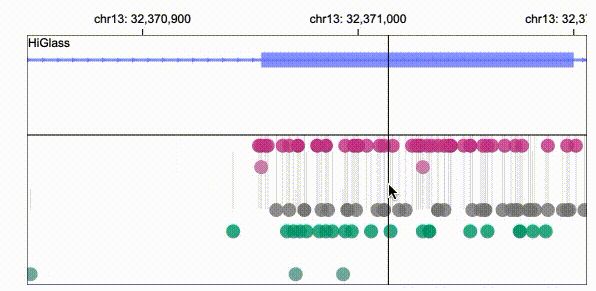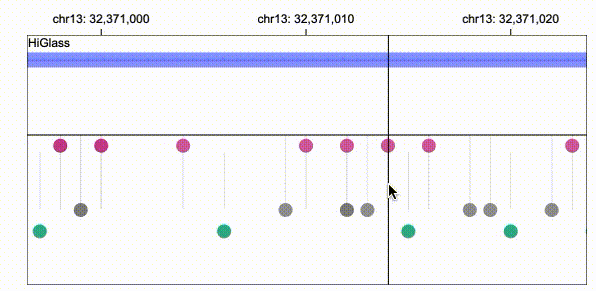
Gosling scales from whole genomes to single nucleotides via semantic zooming that updates visual encodings dynamically and by using the rendering and data access capabilities of our HiGlass genomics visualization framework.
Expressiveness
Gosling is designed to be expressive enough to generate pretty much any visualization of genome-mapped data, which we accomplished by basing the grammar on our taxonomy of (epi)genomics data visualizations.
User Interaction
Gosling has intuitive and effective user interactions built in, including zooming, panning, brushing, and linking. This enables the creation of flexible visualizations that cover a wide range of visual analysis scenarios, like overview + detail views with brushes or comparative views.