Mark
Visual marks (e.g., points, lines, and bars) are the basic graphical elements of a visualization. Note here that we call one visualization a track in Gosling.
The core of constructing a visualization is to bind selected data fields to the visual channels (e.g., size, color, and position) of a chosen mark type.
The mark property of a track is defined by a string that describes the mark type.
{
"tracks":[
/** a track using rect marks */
{
"mark": "rect",
... // other track properties
},
/** a track using line marks */
{
"mark": "line",
... // other track properties
}
],
... // other visualization properties
}
Gosling supports the following primitive mark types: Point, Line,
Area, Bar, Rect, Text, Link, Triangle.
Point
The mark point represents one data point using a circular shape. Visual channels of the circle, such as radius, color, and vertical/horizontal position, are used to represent values of the data point. Popular charts such as scatter plots and bubble charts use point mark.
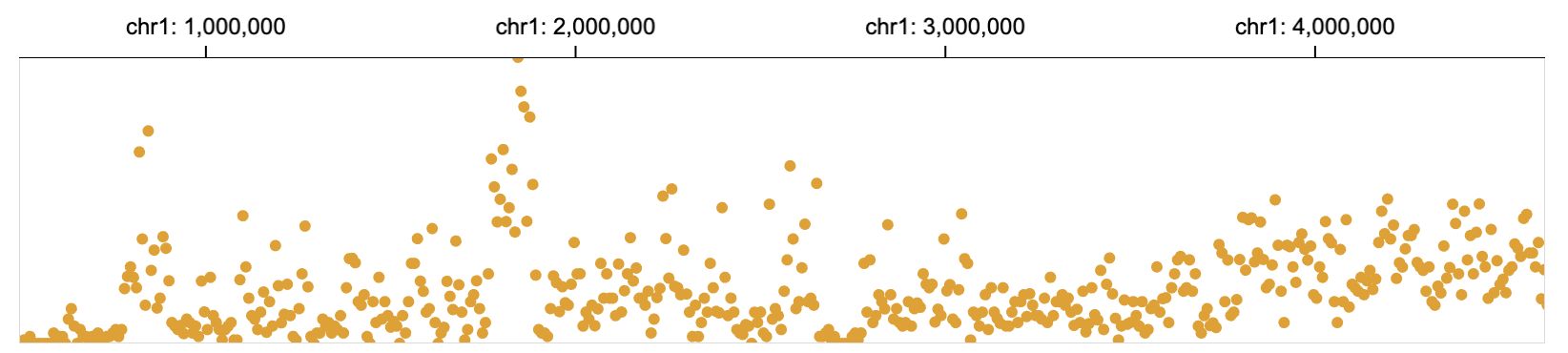
// an example of point marks
{
"tracks":[{
"data": {
"url": ...,
"type": ...
},
// mark type
"mark": "point",
// mark visual channels
"x": {
"field": "position", // data field
"type": "genomic", // type of data field
"axis": "top"
},
"y": {
"field": "peak",
"type": "quantitative"
},
... // other encodings and styles
}]
}
Line
The mark line represents a set of data points using a line that connects these points.
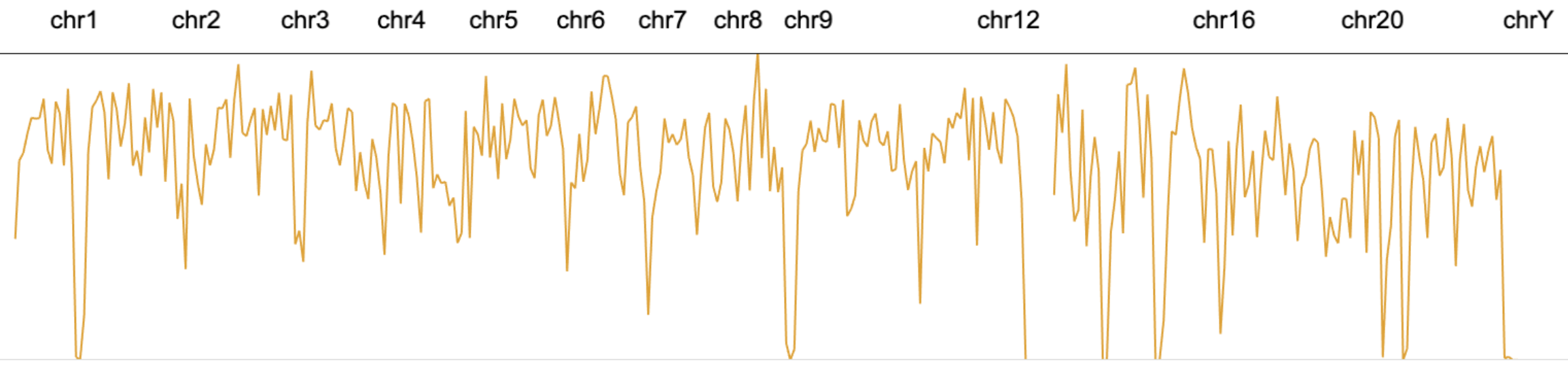
// an example of using line marks
{
"tracks":[{
"data": {
"url": ...,
"type": ...
},
// specify mark type
"mark": "line",
// specify mark visual channels
"x": {
"field": "position", // data field
"type": "genomic", // type of data field
"axis": "top" // position of the x axis
},
"y": {
"field": "peak",
"type": "quantitative"
},
... // other encodings and styles
}]
}
Area
The mark area represents a set of data points as an area shape. The upper edge of the area shape is a line that connects all the points and the bottom edge is the x axis.
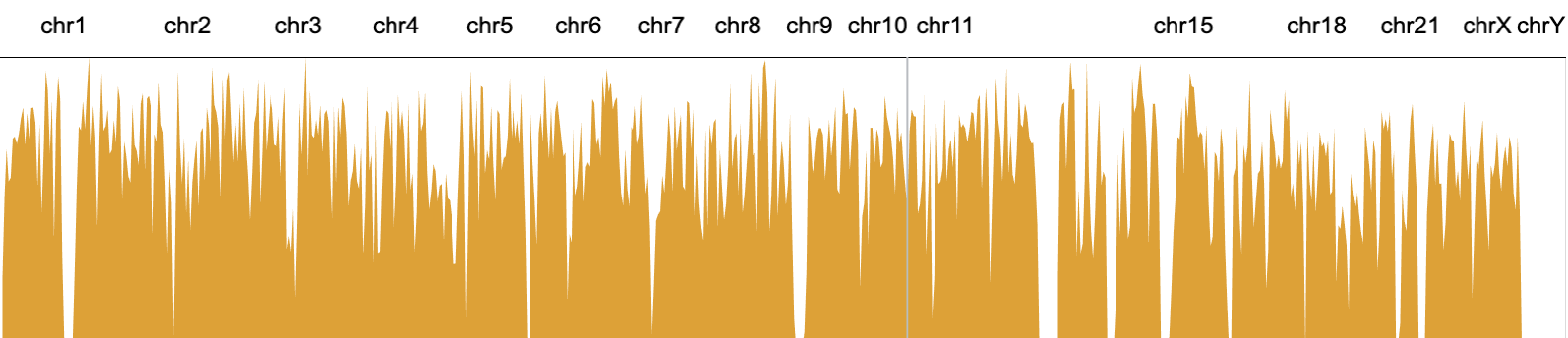
// an example of area marks
{
"tracks":[{
"data": {
"url": ...,
"type": ...
},
// mark type
"mark": "area",
// mark visual channels
"x": {
"field": "position", // data field
"type": "genomic", // type of data field
"axis": "top"
},
"y": {
"field": "peak",
"type": "quantitative"
},
"color": ...,
... // other encodings and styles
}]
}
Bar
The bar mark is designed for drawing bar charts. Each bar shows the value of one data point through its height.
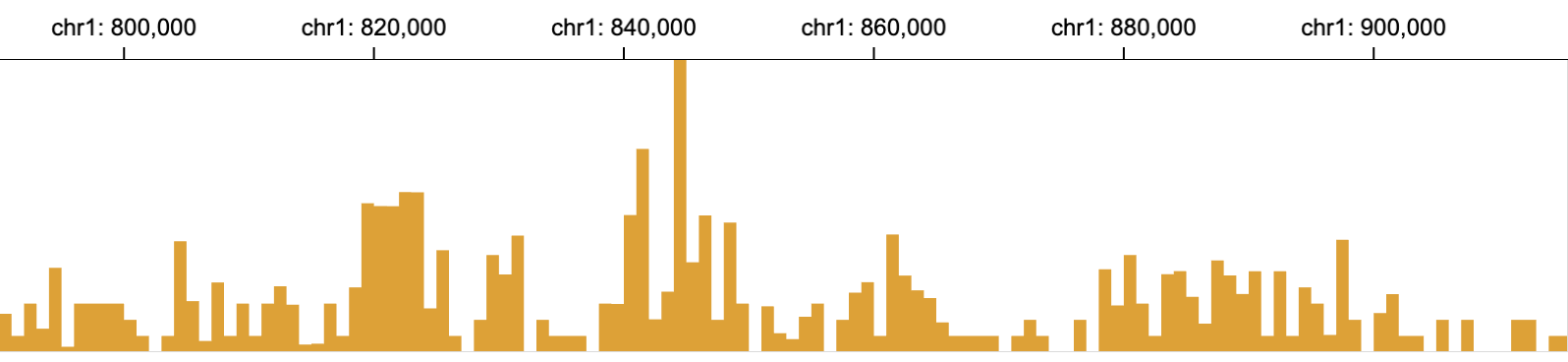
// an example of area marks
{
"tracks":[{
"data": {
"url": ...,
"type": ...
},
// mark type
"mark": "bar",
// mark visual channels
"x": {
"field": "position", // data field
"type": "genomic", // type of data field
"axis": "top"
},
// y indicates the visual encoding of the bar height
"y": {
"field": "peak",
"type": "quantitative"
},
... // other encodings and styles
}]
}
Rect
The rect mark is designed for representing genomic intervals using rectangular shapes. Left and right edge of the rectangle indicate the start and end genomic positions, respectively.

// an example of rect marks
{
"traks":[
{
"data": ...,
// specify the type of mark
"mark": "rect",
// bind the color of each rect mark to the data field: Stain
"color": {
"field": "Stain",
"type": "nominal",,
"domain": ["gneg", "gpos25", "gpos50", "gpos75", "gpos100", "gvar"],
"range": ["white", "#D9D9D9", "#979797", "#636363", "black", "#A0A0F2"]
},
// bind the start position of each rect mark to the data field: chromStart
"x": {
"field": "chromStart",
"type": "genomic",
"axis": "top"
},
// bind the end position of each rect mark to the data field: chromEnd
"xe": {
"field": "chromEnd", "type": "genomic"
},
"size": {
"value": 30 // specify the constant height of each rect mark
}
},
... // other encodings and styles of the rect mark
],
}
The rect mark can also be used for the matrix visualization of genomic interaction data.
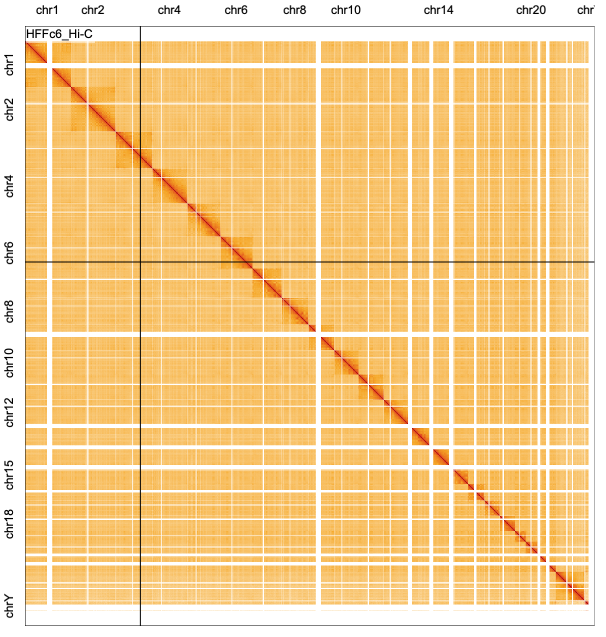
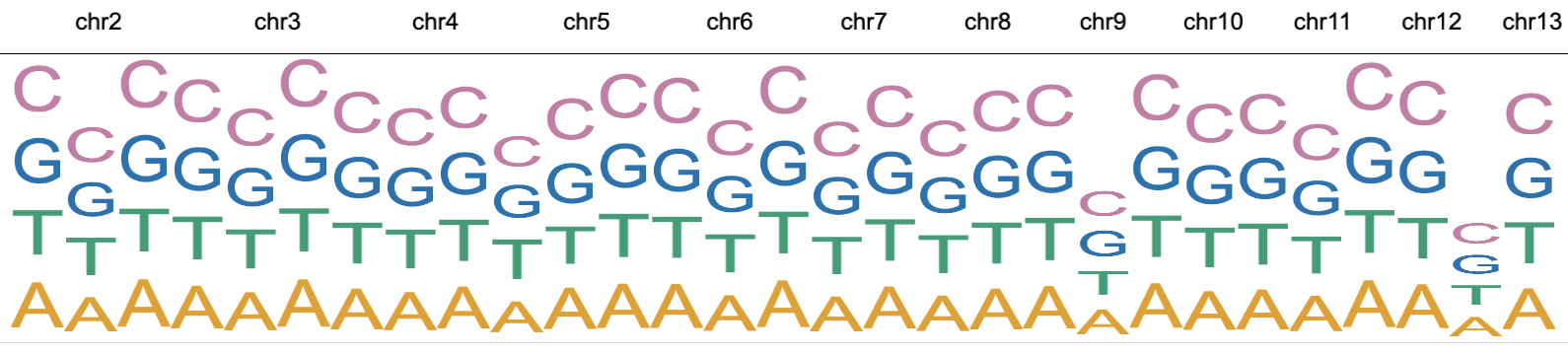
Text
The text mark is designed to display textual labels. For example, gene names and nucleobases can be displayed with text marks.
{
"tracks":[{
"data": ...,
// specify the type of mark
"mark": "text",
// specify styles of the mark
"style": {"textStrokeWidth": 0},
// bind visual channels to corresponding data fields
"x": {"field": "start", "type": "genomic", "axis": "top"},
"xe": {"field": "end", "type": "genomic"},
"y": {"field": "count", "type": "quantitative"},
"color": {
"field": "base",
"type": "nominal",
"domain": ["A", "T", "G", "C"]
},
// specify the text content
"text": {"field": "base", "type": "nominal"}
}]
}
Link
The link mark is designed to show connections between chromosomes.
Gosling supports both "withinLink", links within one track, and "betweenLink", links between different tracks.

{
"tracks": [
{
"data": {...},
"mark": "withinLink", // specify the mark type
// bind visual channels to corresponding data fields
// x and xe indicates the start point of the arc
"x": {
"field": "s1",
"type": "genomic",
"domain": {"chromosome": "1"},
"axis": "top"
},
"xe": {"field": "e1", "type": "genomic"},
// x and xe indicates the end point of the arc
"x1": {
"field": "s2",
"type": "genomic",
"domain": {"chromosome": "1"},
"axis": "top"
},
"x1e": {"field": "e2", "type": "genomic"},
// specify styles of the mark
"stroke": {"value": "steelblue"},
"style": {"circularLink": true}
}
]
}
When connections between points on different chromosomes need to be created, the genomicFieldsToConvert property should be used at the time of loading data.
Currently genomicFieldsToConvert is experimental and only supported in CSV and JSON data.
{
"tracks": [{
"title": "Visualization with data loaded via JSON object",
"data": {
"type": "json",
// convert chromosome fields into genomic fields
"genomicFieldsToConvert": [
{
"chromosomeField": "chrStart",
"genomicFields": ["start1", "end1"]
},
{
"chromosomeField": "chrEnd",
"genomicFields": ["start2", "end2"]
}
],
"values": [
{
"chrStart": "chr1",
"start1": 1221574,
"end1": 1221575,
"chrEnd": "chr13",
"start2": 36001515,
"end2": 36001516
},
{
"chrStart": "chr10",
"start1": 9257519,
"end1": 9257520,
"chrEnd": "chr18",
"start2": 82441834,
"end2": 82441835
}
]
},
"style": {"linkStyle": "elliptical"},
"mark": "withinLink",
// connect chrStart (chr1, chr10) to chrEnd (chr13, chr18)
"x": {"field": "start1", "type": "genomic"},
"xe": {"field": "end2", "type": "genomic"},
"stroke": {"value": "magenta"},
"width": 500,
"height": 180
}]
}
Triangle
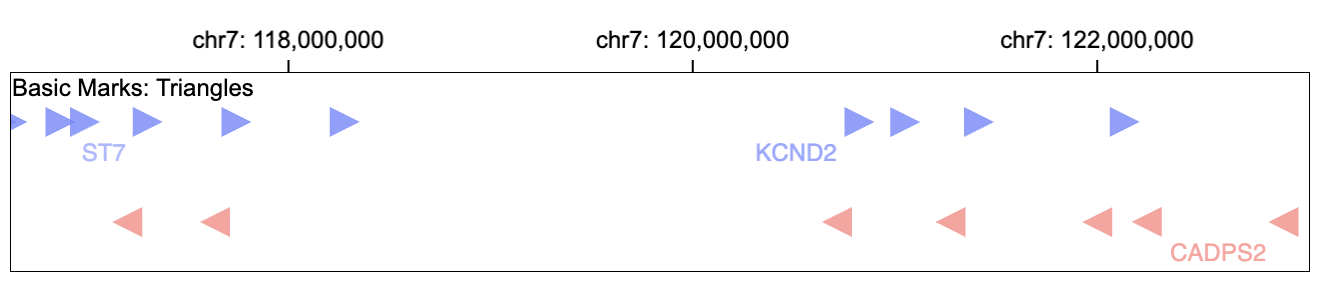
Gosling supports three types of triangle marks: triangleLeft, triangleRight, triangleBottom
{
"tracks": [
{
"data": {...},
"mark": "triangleRight", // specify the mark type
// bind visual channels to corresponding data fields
/** x indicates the position of the mark on a genomic axis */
"x": {
"field": "end",
"type": "genomic",
"axis": "top"
},
/** the size of the triangle */
"size": {"value": 15}
},
{
/** another track */
},
...
]
}